

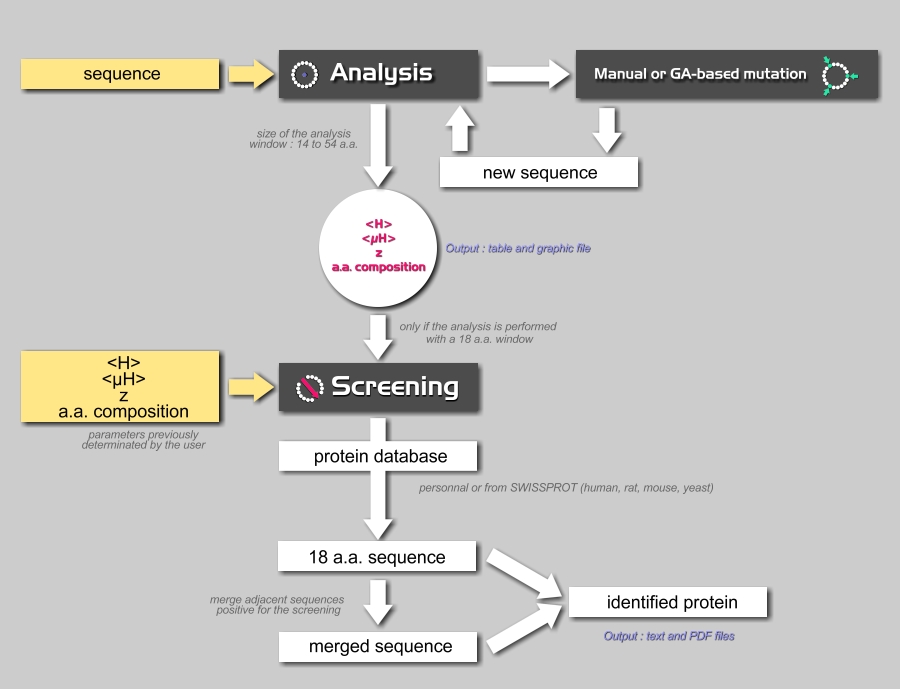
HeliQuest calculates from the amino acid sequence of a helix (α-helix, 3-10 helix, 3-11 helix or π helix) its physicochemical properties and amino acid composition and uses the results to screen any databank in order to identify protein segments possessing similar features.
The server is divided into 2 interconnected modules: the sequence analysis module and the screening module for α-helices and 3-11-helices.
In addition, the mutation module, (available from the sequence analysis module), allows the user to mutate helices manually or automatically using a genetic algorithm, to create analogues with specific properties.
The organization of the server is represented below with the entries and the output files.
Please cite the following reference:
Gautier R., Douguet D., Antonny B. and Drin G. HELIQUEST: a web server to screen sequences with specific α-helical properties. Bioinformatics. 2008 Sep 15;24(18):2101-2.
Comments, feedback, bugs : Please send all remarks to: 
News :2024-03-08 addition of 2 non-natural amino acids (NVA and TIC) to the Analysis module. Update of SwissProt Databases
2021-07-28 Update of SwissProt Databases
2020-03-02 New server and Update of SwissProt databases
2018-10-02 Update of SwissProt databases
2017-11-06 Update of SwissProt databases
2016-06-24 Analysis: modification of the downloadable file (data.txt) with the addition of supplementary data (Moment angle)
2016-03-22 Update of SwissProt databases
2014-12-19 Analysis: possibility to download a data file with the values ÁH, H, z,... for each helix. Screening: Update of SwissProt Databases
2014-03-03 Update of SwissProt Databases (The human database contains human virus sequences)
2012-08-07 Update of SwissProt Databases
2011-07-05 HeliQuest version 2 with new screening module for 3-11 helix and module Analysis for several helix type (α-helix, 3-10 helix, 3-11 helix or π helix)
2011-06-24 Update of SwissProt Databases
2010-12-01 New server, more powerful and fast, Update of the SwissProt Databases
2010-02-02 Update of the SwissProt Databases
2009-04-04 Update of the SwissProt Databases
2008-09-10 Update of the SwissProt Databases
2008-07-01 HeliQuest v1.2 online