

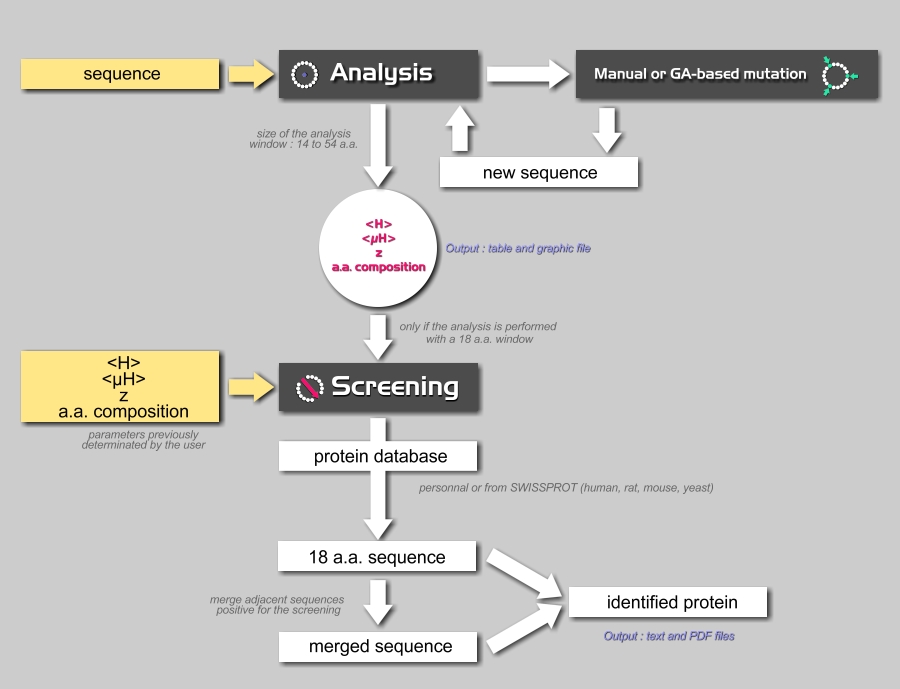
HeliQuest calculates from an helix sequence (α-helix, 3-10 helix, 3-11 helix or π helix) its physicochemical properties and amino acid composition and uses the results to screen any databank in order to identify protein segments possessing similar features.
The server is divided into 2 interconnected modules: the sequence analysis module and the screening module for α-helix sequence and for 3-11-helix sequence.
In addition, the mutation module, (available from the sequence analysis module), allows user to mutate helices manually or automatically by genetic algorithm to create analogues with specific properties.
The organization of the server is represented below with the entries and the output files.
Please cite the following reference:
Gautier R., Douguet D., Antonny B. and Drin G. HELIQUEST: a web server to screen sequences with specific α-helical properties. Bioinformatics. 2008 Sep 15;24(18):2101-2.
Comments, feedback, bugs : Please send all remarks to: 
News :2011-07-05 HeliQuest version 2 with new screening module for 3-11 helix and module Analysis for several helix type (α-helix, 3-10 helix, 3-11 helix or π helix)
2011-06-24 Update of SwissProt Databases
2010-12-01 New server, more powerful and fast, Update of the SwissProt Databases
2010-02-02 Update of the SwissProt Databases
2009-04-04 Update of the SwissProt Databases
2008-09-10 Update of the SwissProt Databases
2008-07-01 HeliQuest v1.2 online